Light microscopy employs visible light to image objects of microscopic dimensions and it is probably the most used research tool in biology. Microscopy has historically been an observational technique. In recent years, however, the development of automated microscopes, digital sensing technologies and novel labeling probes have turned microscopy into a predominantly quantitative technique.
From a systems biology perspective, the cell is the principal element of information integration. Understanding the cell in its spatiotemporal context is the key to unraveling many of the still unknown mechanisms of life and disease. This is a dounting task that requires the study of the cell from many different perspectives (morphological, biochemical, mechanical, electrical) accounting for both the temporal and spatial dimensions. In our group, we use of time-lapse video microscopy to capture the cellular spatiotemporal dynamics.
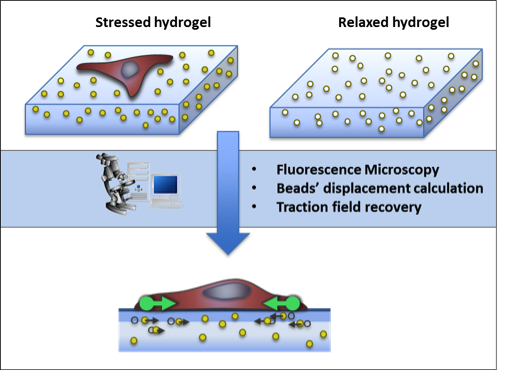
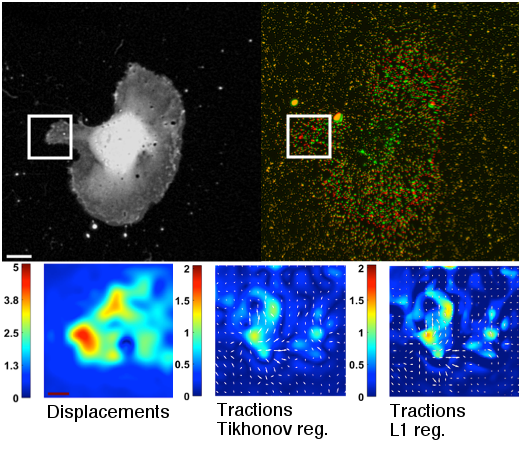
The extraction of biologically relevant information from microscopy imaging modalities requires the use of advance image processing methods. In particular, we focus in the development of algorithms for cell segmentation and tracking, for the characterization of the protrusion dynamics and for the estimation of cell-matrix tensional forces.
Selected Articles
International Journals
A. Jorge-Peñas, A. Izquierdo-Alvarez, R. Aguilar-Cuenca, M. Vicente-Manzanares, J. M. García-Aznar, H. Van Oosterwyck, E. M. de-Juan-Pardo, C. Ortiz-de-Solorzano, A. Muñoz-Barrutia, “Free Form Deformation-Based Image Registration Improves Accuracy of Traction Force Microscopy,” PLoS ONE, 10(12):e0144184, 2015.
C. Ortiz-de-Solorzano, A. Muñoz-Barrutia, E. Meijering, M. Kozubek, "A Morpho-Dynamic Model of the Cell", IEEE Signal Processing Magazine, Special Issue on Quantitative Bioimaging: Signal Processing for Light Microscopy, 21(1):20-29, 2015.
M. Maska, V. Ulman, D. Svoboda, P. Matula, P. Matula, C. Ederra, A. Urbiola, T. España, S. Venkatesan, D. M. W. Balak, T. Bolckova, M. Streitova, C. Carthel, S. Coraluppi, N. Harder, K. Rohr, K. E. G. Magnusson, J. Jalden, H. M. Blau, O. Dzyubachyk, P. Krizek, G. M. Hagen, D. Pastor-Escuredo, D. Jimenez-Carretero, M. J. Ledesma-Carbayo, A. Muñoz-Barrutia, E. Meijering, M. Kozubek, C. Ortiz-de-Solorzano, "A Benchmark for Comparison of Cell Tracking Algorithms", Bioinformatics, 30(11):1609-17, 2014.
T. Pengo, A. Muñoz-Barrutia, I. Zudaire, C. Ortiz-de-Solorzano, "Efficient Blind Spectral Unmixing of Fluorescently Labeled Samples using Multi-Layered Non-Negative Matrix Factorization", PLoS ONE, 8(11):e78504, 2013.
M. Maska, O. Danek, S. Garasa, A. Rouzaut, A. Muñoz-Barrutia, C. Ortiz-de-Solorzano, "Segmentation and Shape Tracking of Whole Fluorescent Cells Based on the Chan-Vese Model", IEEE Transactions on Medical Imaging, 32(6):995-1006, 2013.
Funded Grants
Imaging and measuring cancer cell mechanics. TEC2013-48552-C2. Ministerio de Economía y Competitividad.
Programa de Empleo Juvenil. Comunidad de Madrid.
Development of a gene therapy platform for renal genetic diseases. Desarrollo de una plataforma de terapia génica para enfermedades genéticas renales. Financiado por el Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación / Proyecto RTC-2017-6600-4.

Idiomas